| << Chapter < Page | Chapter >> Page > |
Within 24 hours, the results of the diagnostic test analysis of Alex’s stool sample revealed that it was positive for heat-labile enterotoxin (LT) , heat-stabile enterotoxin (ST) , and colonization factor (CF) , confirming the hospital physician’s suspicion of ETEC . During a follow-up with Alex’s family physician, this physician noted that Alex’s symptoms were not resolving quickly and he was experiencing discomfort that was preventing him from returning to classes. The family physician prescribed Alex a course of ciprofloxacin to resolve his symptoms. Fortunately, the ciprofloxacin resolved Alex’s symptoms within a few days.
Alex likely got his infection from ingesting contaminated food or water. Emerging industrialized countries like Mexico are still developing sanitation practices that prevent the contamination of water with fecal material. Travelers in such countries should avoid the ingestion of undercooked foods, especially meats, seafood, vegetables, and unpasteurized dairy products. They should also avoid use of water that has not been treated; this includes drinking water, ice cubes, and even water used for brushing teeth. Using bottled water for these purposes is a good alternative. Good hygiene (handwashing) can also aid the prevention of an ETEC infection. Alex had not been careful about his food or water consumption, which led to his illness.
Alex’s symptoms were very similar to those of cholera , caused by the gram-negative bacterium Vibrio cholerae , which also produces a toxin similar to ST and LT. At some point in the evolutionary history of ETEC , a nonpathogenic strain of E. coli similar to those typically found in the gut may have acquired the genes encoding the ST and LT toxins from V. cholerae . The fact that the genes encoding those toxins are encoded on extrachromosomal plasmids in ETEC supports the idea that these genes were acquired by E. coli and are likely maintained in bacterial populations through horizontal gene transfer.
Go back to the previous Clinical Focus box.
Viral genomes exhibit significant diversity in structure. Some viruses have genomes that consist of DNA as their genetic material. This DNA may be single stranded, as exemplified by human parvoviruses , or double stranded, as seen in the herpesviruses and poxviruses . Additionally, although all cellular life uses DNA as its genetic material, some viral genomes are made of either single-stranded or double-stranded RNA molecules, as we have discussed. Viral genomes are typically smaller than most bacterial genomes, encoding only a few genes, because they rely on their hosts to carry out many of the functions required for their replication . The diversity of viral genome structures and their implications for viral replication life cycles are discussed in more detail in The Viral Life Cycle .
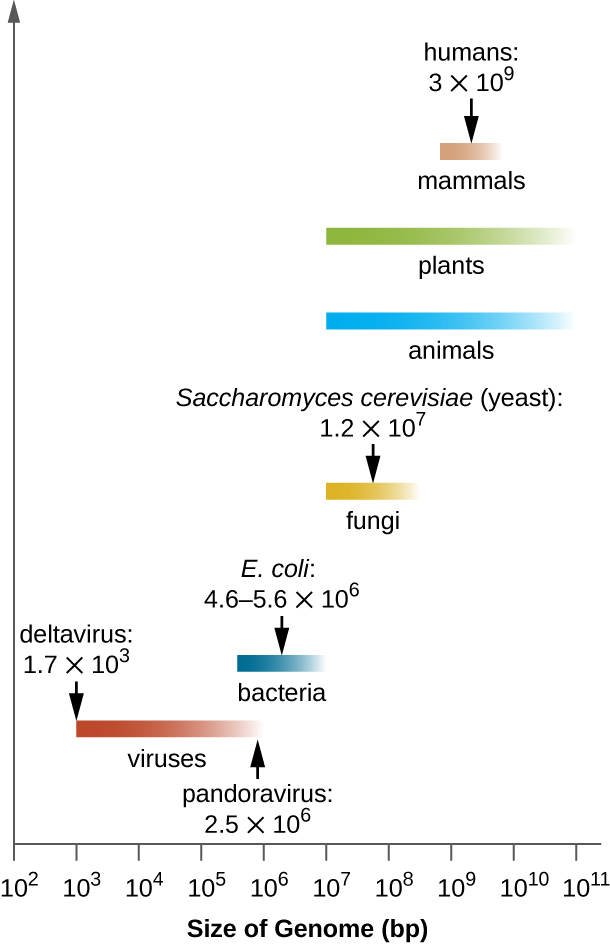
There is great variation in size of genomes among different organisms. Most eukaryotes maintain multiple chromosomes; humans, for example have 23 pairs, giving them 46 chromosomes. Despite being large at 3 billion base pairs, the human genome is far from the largest genome. Plants often maintain very large genomes, up to 150 billion base pairs, and commonly are polyploid, having multiple copies of each chromosome.
The size of bacterial genomes also varies considerably, although they tend to be smaller than eukaryotic genomes ( [link] ). Some bacterial genomes may be as small as only 112,000 base pairs. Often, the size of a bacterium’s genome directly relates to how much the bacterium depends on its host for survival. When a bacterium relies on the host cell to carry out certain functions, it loses the genes encoding the abilities to carry out those functions itself. These types of bacterial endosymbionts are reminiscent of the prokaryotic origins of mitochondria and chloroplasts.
From a clinical perspective, obligate intracellular pathogens also tend to have small genomes (some around 1 million base pairs). Because host cells supply most of their nutrients, they tend to have a reduced number of genes encoding metabolic functions. Due to their small sizes, the genomes of organisms like Mycoplasma genitalium (580,000 base pairs), Chlamydia trachomatis (1.0 million), Rickettsia prowazekii (1.1 million), and Treponema pallidum (1.1 million) were some of the earlier bacterial genomes sequenced. Respectively, these pathogens cause urethritis and pelvic inflammation, chlamydia, typhus, and syphilis.
Whereas obligate intracellular pathogens have unusually small genomes, other bacteria with a great variety of metabolic and enzymatic capabilities have unusually large bacterial genomes. Pseudomonas aeruginosa , for example, is a bacterium commonly found in the environment and is able to grow on a wide range of substrates. Its genome contains 6.3 million base pairs, giving it a high metabolic ability and the ability to produce virulence factors that cause several types of opportunistic infections .
Interestingly, there has been significant variability in genome size in viruses as well, ranging from 3,500 base pairs to 2.5 million base pairs, significantly exceeding the size of many bacterial genomes. The great variation observed in viral genome sizes further contributes to the great diversity of viral genome characteristics already discussed.
Visit the genome database of the National Center for Biotechnology Information (NCBI) to see the genomes that have been sequenced and their sizes.
Within an organism, phenotypes may change while genotypes remain constant.
True
Plasmids are typically transferred among members of a bacterial community by ________ gene transfer.
horizontal
What are some differences in chromosomal structures between prokaryotes and eukaryotes?
How do prokaryotes and eukaryotes manage to fit their lengthy DNA inside of cells? Why is this necessary?
What are some functions of noncoding DNA?
In the chromatin of eukaryotic cells, which regions of the chromosome would you expect to be more compact: the regions that contain genes being actively copied into RNA or those that contain inactive genes?

Notification Switch
Would you like to follow the 'Microbiology' conversation and receive update notifications?