| << Chapter < Page | Chapter >> Page > |
Several errors in sex chromosome number have been characterized. Individuals with three X chromosomes, called triplo-X, are phenotypically female but express developmental delays and reduced fertility. The XXY genotype, corresponding to one type of Klinefelter syndrome, corresponds to phenotypically male individuals with small testes, enlarged breasts, and reduced body hair. More complex types of Klinefelter syndrome exist in which the individual has as many as five X chromosomes. In all types, every X chromosome except one undergoes inactivation to compensate for the excess genetic dosage. This can be seen as several Barr bodies in each cell nucleus. Turner syndrome, characterized as an X0 genotype (i.e., only a single sex chromosome), corresponds to a phenotypically female individual with short stature, webbed skin in the neck region, hearing and cardiac impairments, and sterility.
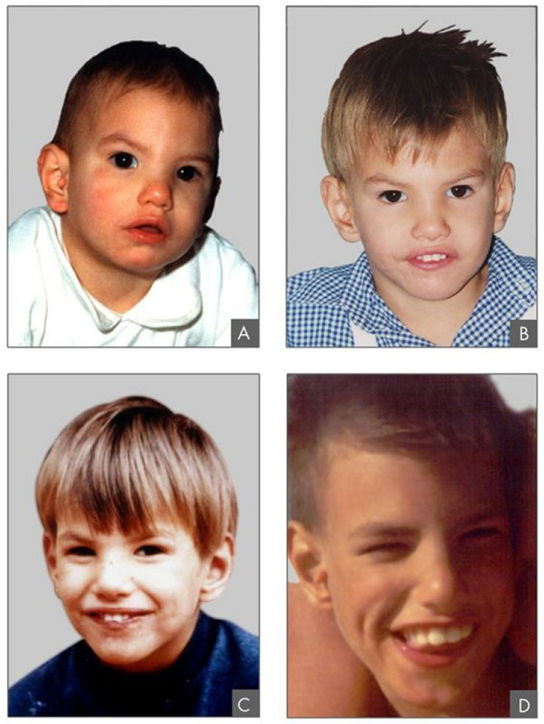
In addition to the loss or gain of an entire chromosome, a chromosomal segment may be duplicated or lost. Duplications and deletions often produce offspring that survive but exhibit physical and mental abnormalities. Duplicated chromosomal segments may fuse to existing chromosomes or may be free in the nucleus. Cri-du-chat (from the French for “cry of the cat”) is a syndrome associated with nervous system abnormalities and identifiable physical features that result from a deletion of most of 5p (the small arm of chromosome 5) ( [link] ). Infants with this genotype emit a characteristic high-pitched cry on which the disorder’s name is based.
Cytologists have characterized numerous structural rearrangements in chromosomes, but chromosome inversions and translocations are the most common. Both are identified during meiosis by the adaptive pairing of rearranged chromosomes with their former homologs to maintain appropriate gene alignment. If the genes carried on two homologs are not oriented correctly, a recombination event could result in the loss of genes from one chromosome and the gain of genes on the other. This would produce aneuploid gametes.
A chromosome inversion is the detachment, 180° rotation, and reinsertion of part of a chromosome. Inversions may occur in nature as a result of mechanical shear, or from the action of transposable elements (special DNA sequences capable of facilitating the rearrangement of chromosome segments with the help of enzymes that cut and paste DNA sequences). Unless they disrupt a gene sequence, inversions only change the orientation of genes and are likely to have more mild effects than aneuploid errors. However, altered gene orientation can result in functional changes because regulators of gene expression could be moved out of position with respect to their targets, causing aberrant levels of gene products.
An inversion can be pericentric and include the centromere, or paracentric and occur outside of the centromere ( [link] ). A pericentric inversion that is asymmetric about the centromere can change the relative lengths of the chromosome arms, making these inversions easily identifiable.

Notification Switch
Would you like to follow the 'Open genetics' conversation and receive update notifications?