| << Chapter < Page | Chapter >> Page > |
Like prokaryotic cells, the transcription of genes in eukaryotes requires the actions of an RNA polymerase to bind to a sequence upstream of a gene to initiate transcription. However, unlike prokaryotic cells, the eukaryotic RNA polymerase requires other proteins, or transcription factors, to facilitate transcription initiation. Transcription factors are proteins that bind to the promoter sequence and other regulatory sequences to control the transcription of the target gene. RNA polymerase by itself cannot initiate transcription in eukaryotic cells. Transcription factors must bind to the promoter region first and recruit RNA polymerase to the site for transcription to be established.
View the process of transcription—the making of RNA from a DNA template—at this site .
Genes are organized to make the control of gene expression easier. The promoter region is immediately upstream of the coding sequence. This region can be short (only a few nucleotides in length) or quite long (hundreds of nucleotides long). The longer the promoter, the more available space for proteins to bind. This also adds more control to the transcription process. The length of the promoter is gene-specific and can differ dramatically between genes. Consequently, the level of control of gene expression can also differ quite dramatically between genes. The purpose of the promoter is to bind transcription factors that control the initiation of transcription.
Within the promoter region, just upstream of the transcriptional start site, resides the TATA box. This box is simply a repeat of thymine and adenine dinucleotides (literally, TATA repeats). RNA polymerase binds to the transcription initiation complex, allowing transcription to occur. To initiate transcription, a transcription factor (TFIID) is the first to bind to the TATA box. Binding of TFIID recruits other transcription factors, including TFIIB, TFIIE, TFIIF, and TFIIH to the TATA box. Once this complex is assembled, RNA polymerase can bind to its upstream sequence. When bound along with the transcription factors, RNA polymerase is phosphorylated. This releases part of the protein from the DNA to activate the transcription initiation complex and places RNA polymerase in the correct orientation to begin transcription; DNA-bending protein brings the enhancer, which can be quite a distance from the gene, in contact with transcription factors and mediator proteins ( [link] ).
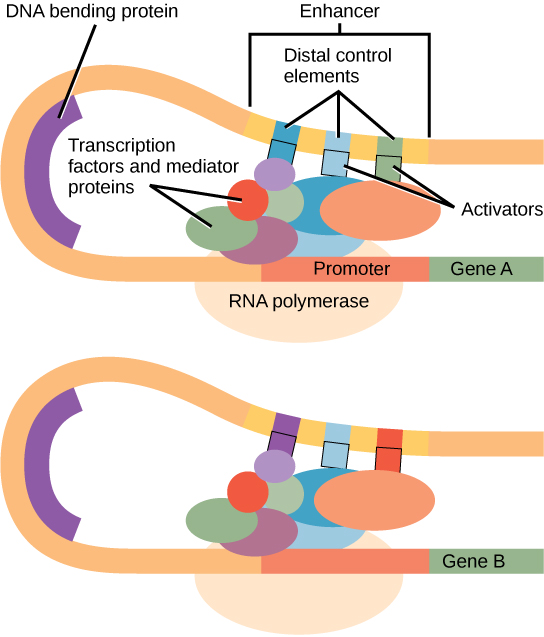
In addition to the general transcription factors, other transcription factors can bind to the promoter to regulate gene transcription. These transcription factors bind to the promoters of a specific set of genes. They are not general transcription factors that bind to every promoter complex, but are recruited to a specific sequence on the promoter of a specific gene. There are hundreds of transcription factors in a cell that each bind specifically to a particular DNA sequence motif. When transcription factors bind to the promoter just upstream of the encoded gene, it is referred to as a cis -acting element , because it is on the same chromosome just next to the gene. The region that a particular transcription factor binds to is called the transcription factor binding site . Transcription factors respond to environmental stimuli that cause the proteins to find their binding sites and initiate transcription of the gene that is needed.
In some eukaryotic genes, there are regions that help increase or enhance transcription. These regions, called enhancers , are not necessarily close to the genes they enhance. They can be located upstream of a gene, within the coding region of the gene, downstream of a gene, or may be thousands of nucleotides away.
Enhancer regions are binding sequences, or sites, for transcription factors. When a DNA-bending protein binds, the shape of the DNA changes ( [link] ). This shape change allows for the interaction of the activators bound to the enhancers with the transcription factors bound to the promoter region and the RNA polymerase. Whereas DNA is generally depicted as a straight line in two dimensions, it is actually a three-dimensional object. Therefore, a nucleotide sequence thousands of nucleotides away can fold over and interact with a specific promoter.
Like prokaryotic cells, eukaryotic cells also have mechanisms to prevent transcription. Transcriptional repressors can bind to promoter or enhancer regions and block transcription. Like the transcriptional activators, repressors respond to external stimuli to prevent the binding of activating transcription factors.
To start transcription, general transcription factors, such as TFIID, TFIIH, and others, must first bind to the TATA box and recruit RNA polymerase to that location. The binding of additional regulatory transcription factors to cis -acting elements will either increase or prevent transcription. In addition to promoter sequences, enhancer regions help augment transcription. Enhancers can be upstream, downstream, within a gene itself, or on other chromosomes. Transcription factors bind to enhancer regions to increase or prevent transcription.

Notification Switch
Would you like to follow the 'Gene expression (gpc)' conversation and receive update notifications?