| << Chapter < Page | Chapter >> Page > |
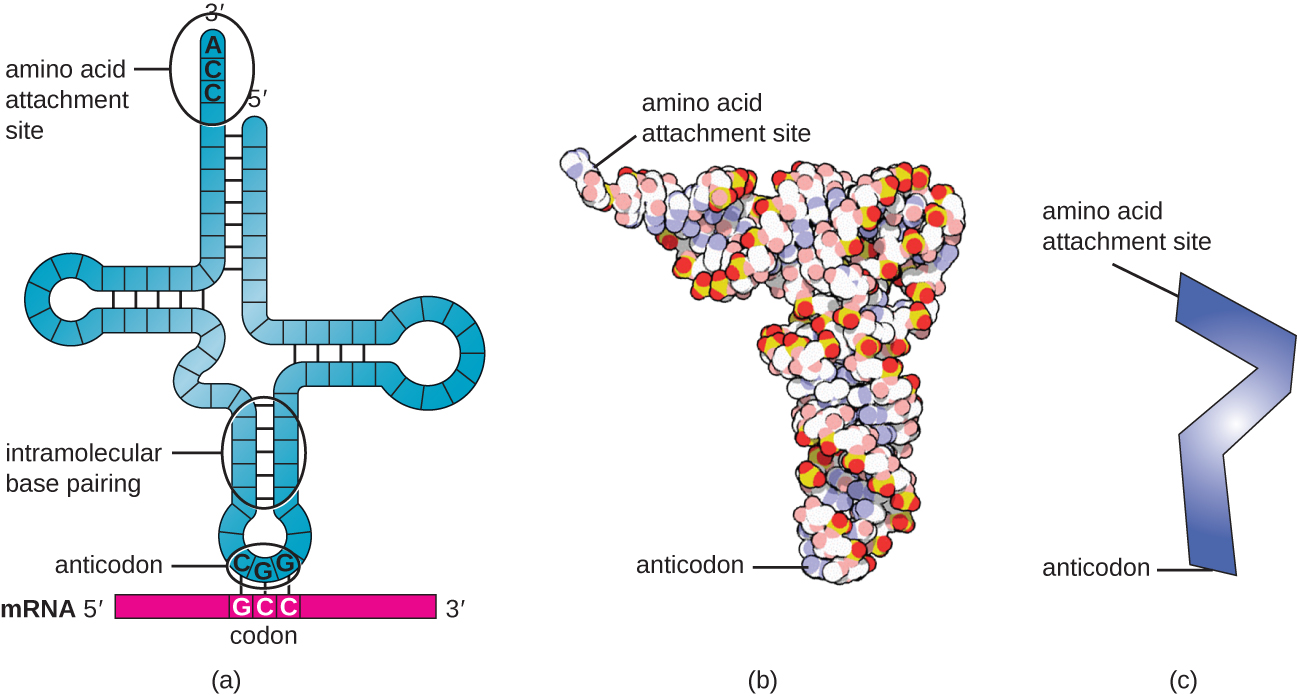
Transfer RNAs (tRNAs) are structural RNA molecules and, depending on the species, many different types of tRNAs exist in the cytoplasm. Bacterial species typically have between 60 and 90 types. Serving as adaptors, each tRNA type binds to a specific codon on the mRNA template and adds the corresponding amino acid to the polypeptide chain. Therefore, tRNAs are the molecules that actually “translate” the language of RNA into the language of proteins. As the adaptor molecules of translation, it is surprising that tRNAs can fit so much specificity into such a small package. The tRNA molecule interacts with three factors: aminoacyl tRNA synthetases, ribosomes, and mRNA.
Mature tRNAs take on a three-dimensional structure when complementary bases exposed in the single-stranded RNA molecule hydrogen bond with each other ( [link] ). This shape positions the amino-acid binding site, called the CCA amino acid binding end , which is a cytosine-cytosine-adenine sequence at the 3’ end of the tRNA, and the anticodon at the other end. The anticodon is a three-nucleotide sequence that bonds with an mRNA codon through complementary base pairing.
An amino acid is added to the end of a tRNA molecule through the process of tRNA “charging,” during which each tRNA molecule is linked to its correct or cognate amino acid by a group of enzymes called aminoacyl tRNA synthetase s. At least one type of aminoacyl tRNA synthetase exists for each of the 20 amino acids. During this process, the amino acid is first activated by the addition of adenosine monophosphate (AMP) and then transferred to the tRNA, making it a charged tRNA , and AMP is released.
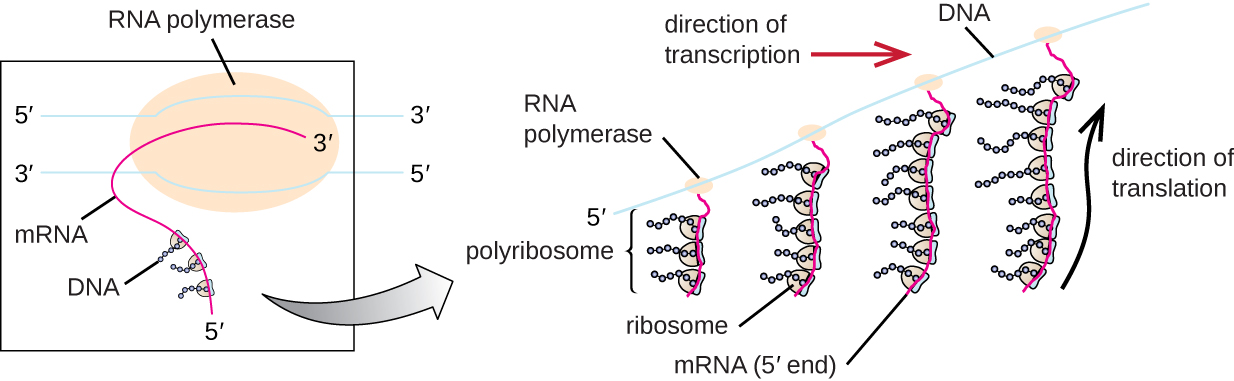
Translation is similar in prokaryotes and eukaryotes. Here we will explore how translation occurs in E. coli , a representative prokaryote, and specify any differences between bacterial and eukaryotic translation.
The initiation of protein synthesis begins with the formation of an initiation complex. In E. coli , this complex involves the small 30S ribosome, the mRNA template, three initiation factors that help the ribosome assemble correctly, guanosine triphosphate (GTP) that acts as an energy source, and a special initiator tRNA carrying N -formyl-methionine (fMet-tRNA fMet ) ( [link] ). The initiator tRNA interacts with the start codon AUG of the mRNA and carries a formylated methionine (fMet). Because of its involvement in initiation, fMet is inserted at the beginning (N terminus) of every polypeptide chain synthesized by E. coli . In E. coli mRNA, a leader sequence upstream of the first AUG codon, called the Shine-Dalgarno sequence (also known as the ribosomal binding site AGGAGG), interacts through complementary base pairing with the rRNA molecules that compose the ribosome. This interaction anchors the 30S ribosomal subunit at the correct location on the mRNA template. At this point, the 50S ribosomal subunit then binds to the initiation complex, forming an intact ribosome.

Notification Switch
Would you like to follow the 'Microbiology' conversation and receive update notifications?