| << Chapter < Page | Chapter >> Page > |
Visualize how mRNA splicing happens by watching the process in action in this video .
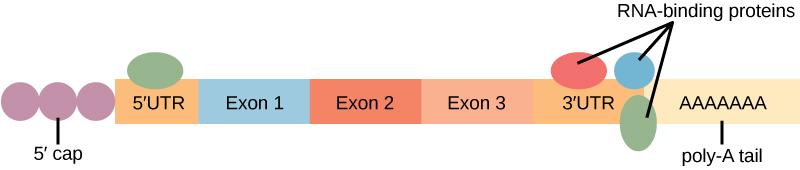
Before the mRNA leaves the nucleus, it is given two protective "caps" that prevent the end of the strand from degrading during its journey. The 5' cap , which is placed on the 5' end of the mRNA, is usually composed of a methylated guanosine triphosphate molecule (GTP). The poly-A tail , which is attached to the 3' end, is usually composed of a series of adenine nucleotides. Once the RNA is transported to the cytoplasm, the length of time that the RNA resides there can be controlled. Each RNA molecule has a defined lifespan and decays at a specific rate. This rate of decay can influence how much protein is in the cell. If the decay rate is increased, the RNA will not exist in the cytoplasm as long, shortening the time for translation to occur. Conversely, if the rate of decay is decreased, the RNA molecule will reside in the cytoplasm longer and more protein can be translated. This rate of decay is referred to as the RNA stability. If the RNA is stable, it will be detected for longer periods of time in the cytoplasm.
Binding of proteins to the RNA can influence its stability. Proteins, called RNA-binding proteins , or RBPs, can bind to the regions of the RNA just upstream or downstream of the protein-coding region. These regions in the RNA that are not translated into protein are called the untranslated regions , or UTRs. They are not introns (those have been removed in the nucleus). Rather, these are regions that regulate mRNA localization, stability, and protein translation. The region just before the protein-coding region is called the 5' UTR , whereas the region after the coding region is called the 3' UTR ( [link] ). The binding of RBPs to these regions can increase or decrease the stability of an RNA molecule, depending on the specific RBP that binds.
In addition to RBPs that bind to and control (increase or decrease) RNA stability, other elements called microRNAs can bind to the RNA molecule. These microRNAs , or miRNAs, are short RNA molecules that are only 21–24 nucleotides in length. The miRNAs are made in the nucleus as longer pre-miRNAs. These pre-miRNAs are chopped into mature miRNAs by a protein called dicer . Like transcription factors and RBPs, mature miRNAs recognize a specific sequence and bind to the RNA; however, miRNAs also associate with a ribonucleoprotein complex called the RNA-induced silencing complex (RISC) . RISC binds along with the miRNA to degrade the target mRNA. Together, miRNAs and the RISC complex rapidly destroy the RNA molecule.
Post-transcriptional control can occur at any stage after transcription, including RNA splicing, nuclear shuttling, and RNA stability. Once RNA is transcribed, it must be processed to create a mature RNA that is ready to be translated. This involves the removal of introns that do not code for protein. Spliceosomes bind to the signals that mark the exon/intron border to remove the introns and ligate the exons together. Once this occurs, the RNA is mature and can be translated. RNA is created and spliced in the nucleus, but needs to be transported to the cytoplasm to be translated. RNA is transported to the cytoplasm through the nuclear pore complex. Once the RNA is in the cytoplasm, the length of time it resides there before being degraded, called RNA stability, can also be altered to control the overall amount of protein that is synthesized. The RNA stability can be increased, leading to longer residency time in the cytoplasm, or decreased, leading to shortened time and less protein synthesis. RNA stability is controlled by RNA-binding proteins (RPBs) and microRNAs (miRNAs). These RPBs and miRNAs bind to the 5' UTR or the 3' UTR of the RNA to increase or decrease RNA stability. Depending on the RBP, the stability can be increased or decreased significantly; however, miRNAs always decrease stability and promote decay.

Notification Switch
Would you like to follow the 'Biology' conversation and receive update notifications?